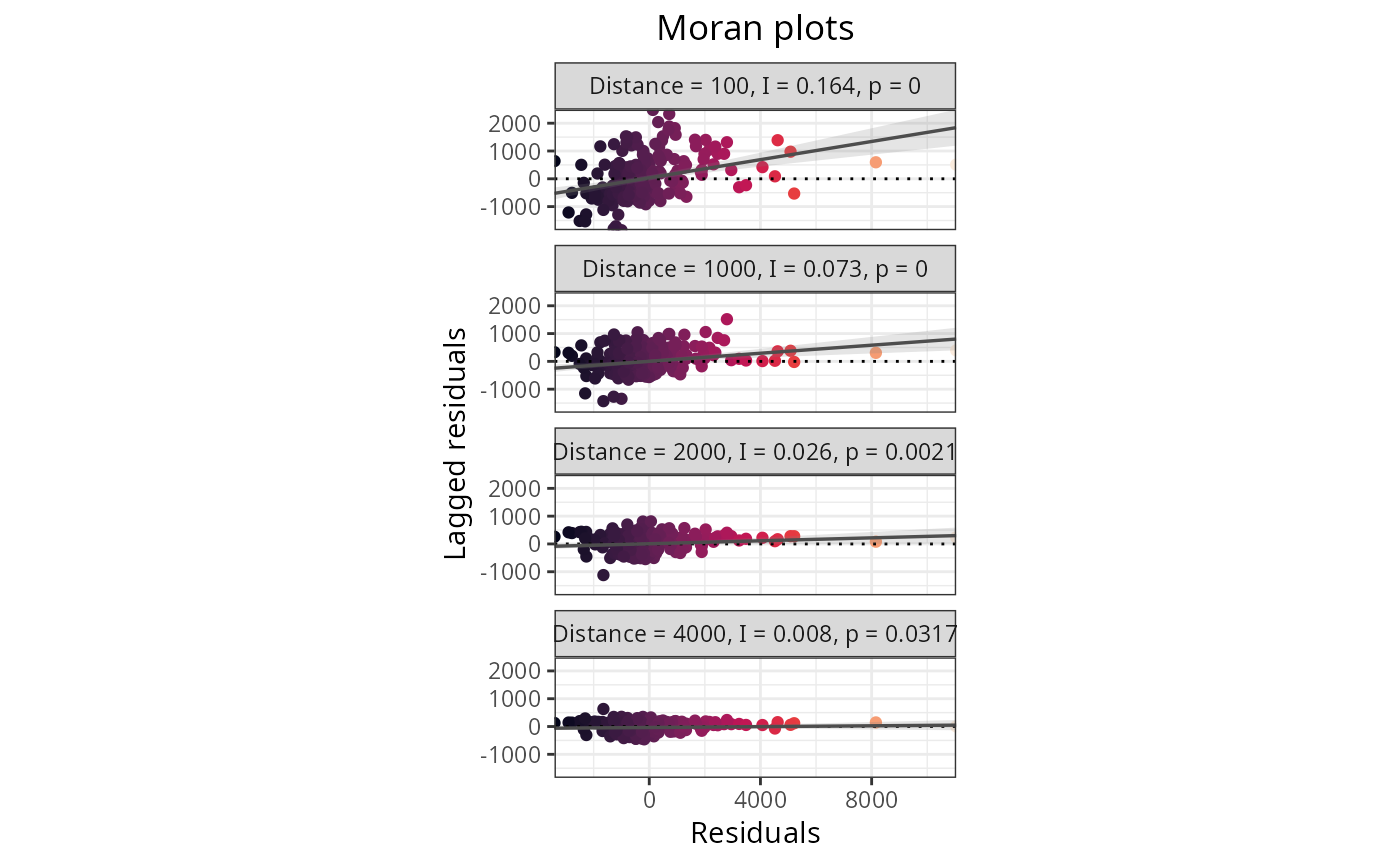
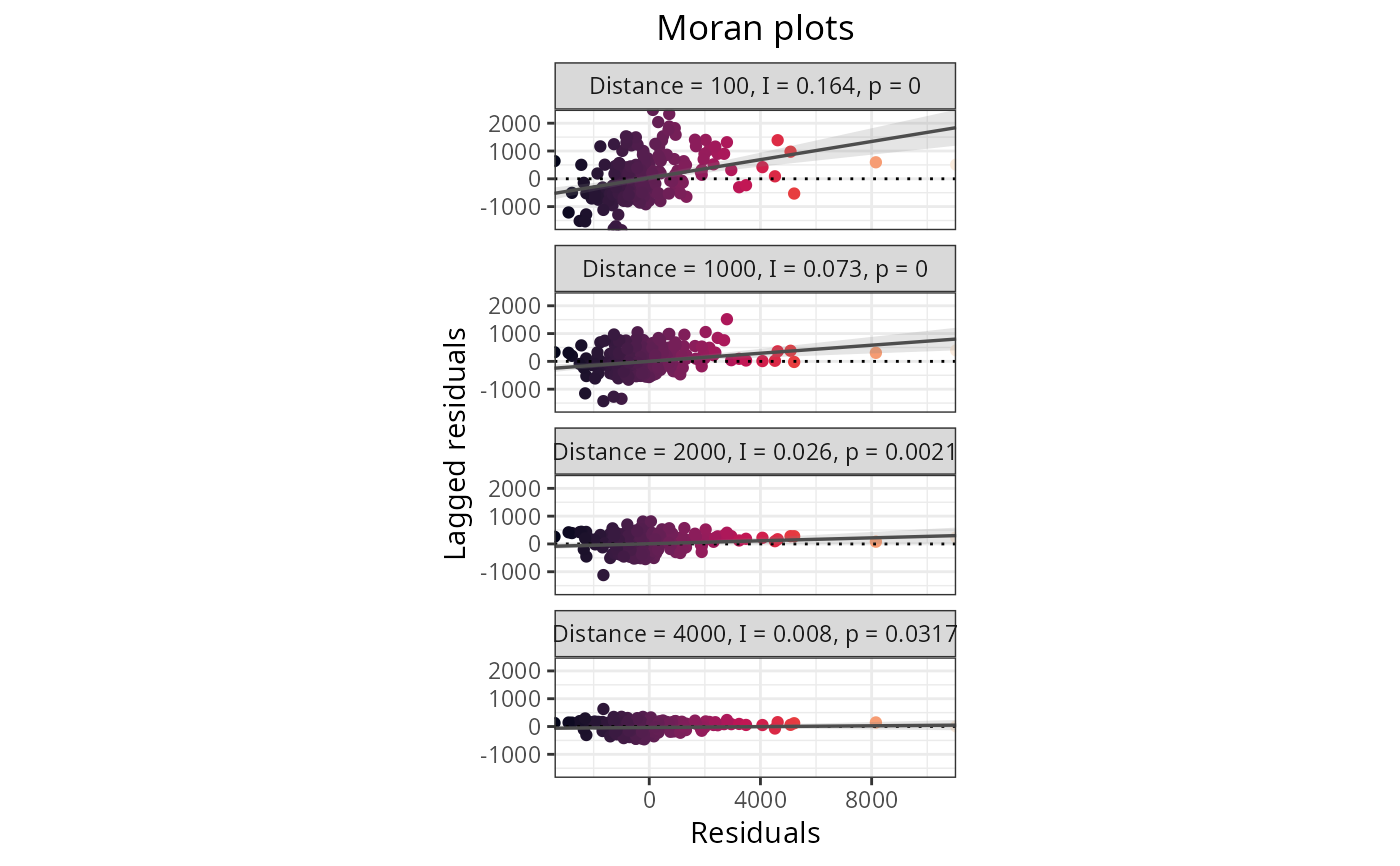
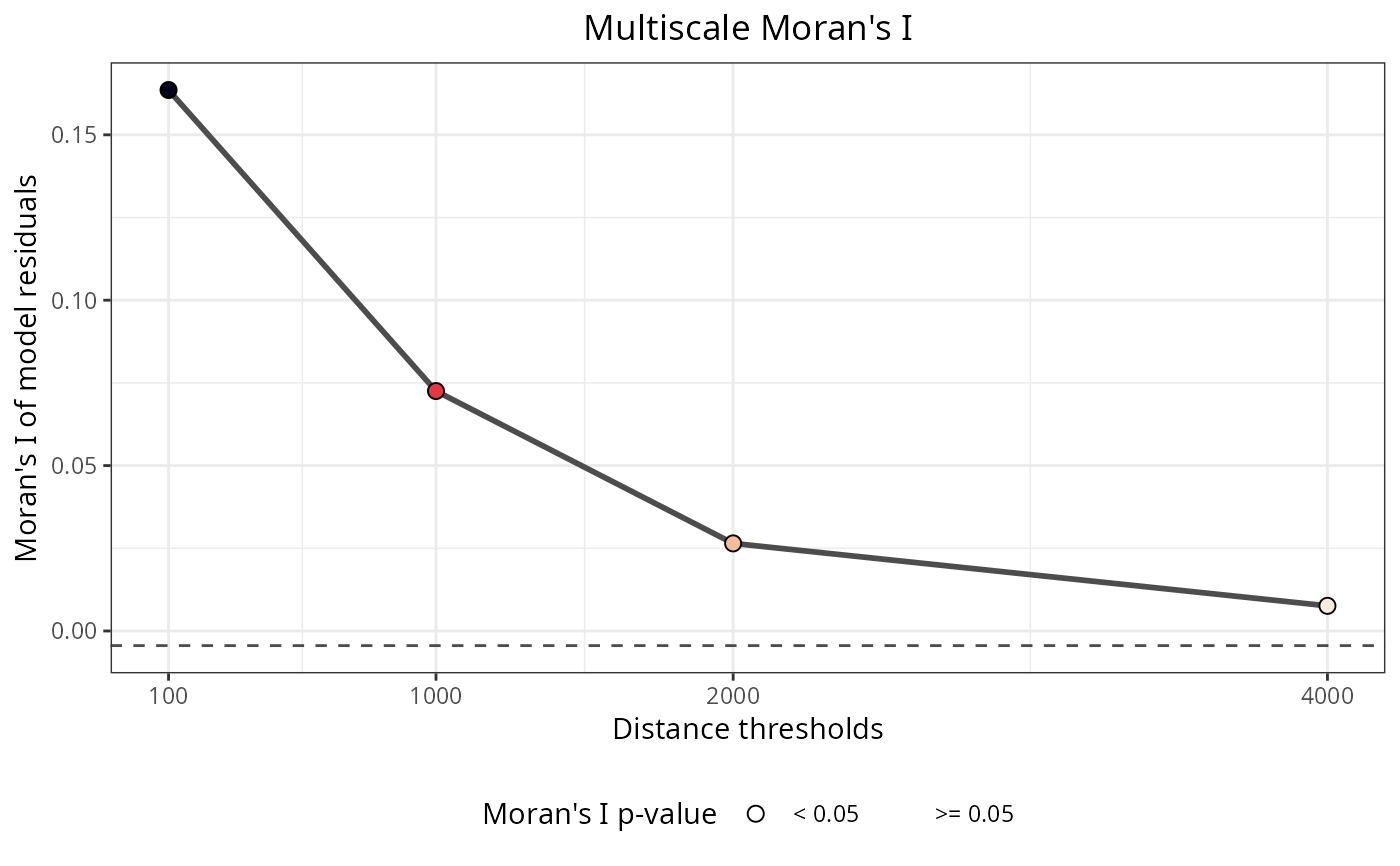
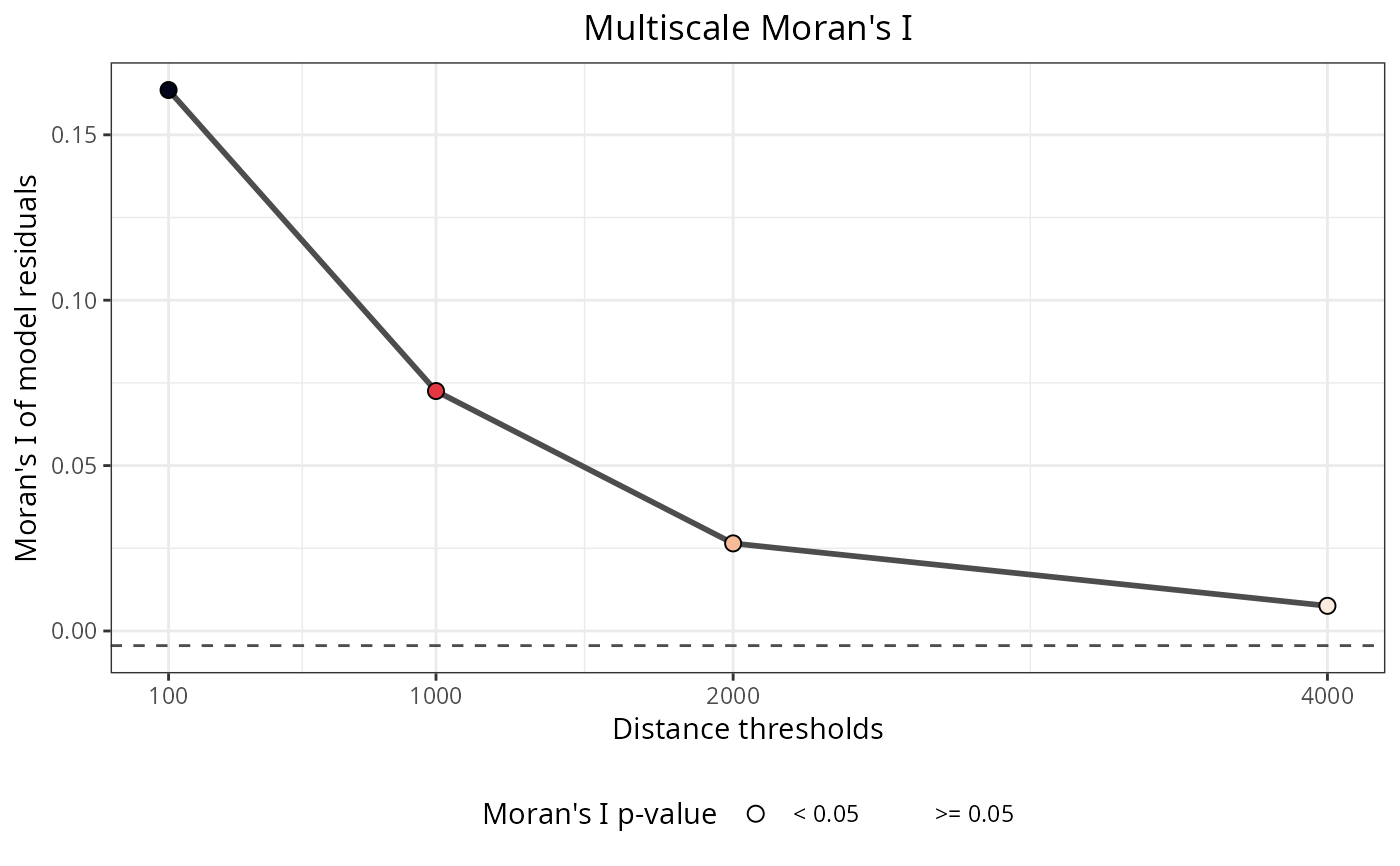
Plots the results of spatial autocorrelation tests for a variety of functions within the package. The x axis represents the Moran's I estimate, the y axis contains the values of the distance thresholds, the dot sizes represent the p-values of the Moran's I estimate, and the red dashed line represents the theoretical null value of the Moran's I estimate.
Usage
plot_moran(
model,
point.color = viridis::viridis(
100,
option = "F",
direction = -1
),
line.color = "gray30",
option = 1,
ncol = 1,
verbose = TRUE
)Arguments
- model
A model fitted with
rf(),rf_repeat(), orrf_spatial(), or a data frame generated bymoran(). Default:NULL- point.color
Colors of the plotted points. Can be a single color name (e.g. "red4"), a character vector with hexadecimal codes (e.g. "#440154FF" "#21908CFF" "#FDE725FF"), or function generating a palette (e.g.
viridis::viridis(100)). Default:viridis::viridis(100, option = "F")- line.color
Character string, color of the line produced by
ggplot2::geom_smooth(). Default:"gray30"- option
Integer, type of plot. If
1(default) a line plot with Moran's I and p-values across distance thresholds is returned. If2, scatterplots of residuals versus lagged residuals per distance threshold and their corresponding slopes are returned. In models fitted withrf_repeat(), the residuals and lags of the residuals are computed from the median residuals across repetitions. Option2is disabled ifxis a data frame generated bymoran().- ncol
Number of columns of the plot. Only relevant when
option = 2. Argumentncolof wrap_plots.- verbose
Logical, if
TRUE, the resulting plot is printed, Default:TRUE
Examples
data(plants_rf)
plot_moran(plants_rf)
 plot_moran(plants_rf, option = 2)
plot_moran(plants_rf, option = 2)